Mathematical models of signaling pathways and screening approaches
Upon the identification of novel, discrete signaling networks, we can employ more directed mathematical methods to dissect the signaling pathway and the contributions from its distinct nodes. These powerful computational and mathematical methods are applicable to a wide range of model systems, from cells to flies to humans. The greatest burden, yet most enjoyable, is to know how to ask the correct question to be analyzed.
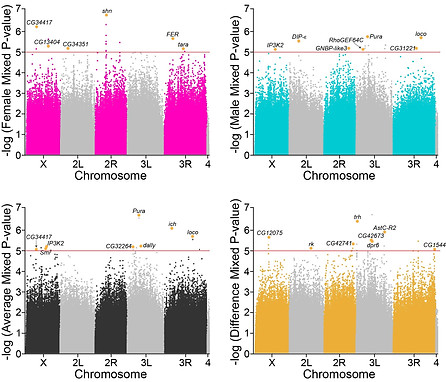
For example, we used data collected in mouse cell lines to model how translational control mechanisms regulate influenza virus replication (Goodman and Tanner et al., 2011). More recently, we used the Drosophila Genetic Reference Panel to analyze how ~4.6 million gene variants contributed to Coxiella burnetii pathogenesis and uncovered 15 genes that regulate pathogenesis (Guzman and Howard et al, 2021).